Reproducible Research
Here we present some general best practices for open and reproducible science. Researchers in the group are strongly encouraged to adhere to these practices for all their data analysis projects.
Project-Oriented Workflow
Working directory
When working on a self-contained project as the one described before, the working directory must be set to the project’s root folder.
Ideally, this is achieved via development workflow and tooling, not by placing calls to setwd() with absolute paths into your scripts. Using an Integrated Development Environment (IDE) that supports a project-based workflow is strongly recommended. This eliminates the tension between your development convenience and the portability of the code. More details in the next section.
When opening an RStudio project, RStudio will set the working directory to the root of the project automatically.
Fresh R sessions
Every time you open a project, it should start in a fresh R session to avoid any interactions with code you might have run beforehand.
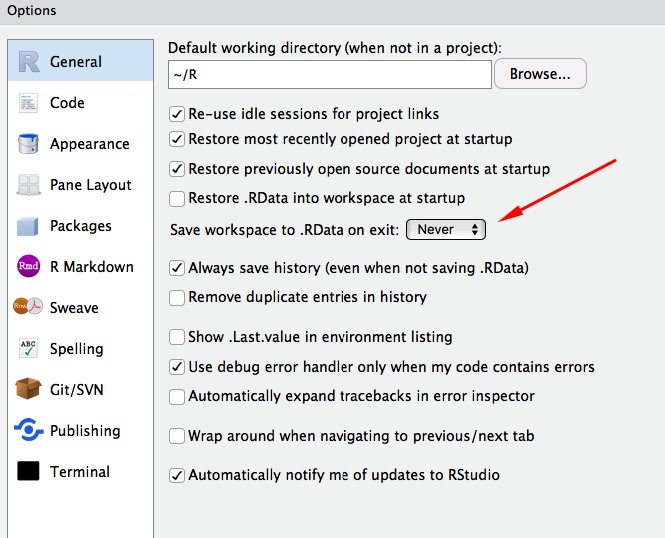
A consequence of this approach is that you should never save .RData when you quit R or load .RData when you start R.
This can be disabled by default in RStudio’s Global Options:
Or if you run R from the shell use R --no-save --no-restore-data. You may even set this as the default by putting this line in your .bash_profile:
alias R='R --no-save --no-restore-data'You should restart R very often an re-run your under-developement script from the top.
Note that rm(list = ls()) does not make a fresh R session since this will not detach packages.
File path discipline
In general, no absolute paths should appear in your scripts. Absolute paths are specific to your machine. That is why setting your working directory by baking setwd("C:\Users\sergio\path\that\only\I\have") into your scripts will not work in other computer. If you change computers or someone else tries to run your analysis on her machine, the code will not work.
All paths must be relative to the project’s root folder. That is why we need to include all the necessary files inside our project’s folder. With this setup, we can move the project folder to different folders in our computer, or even to different computers entirely. Note that relative paths are necessary but not sufficient for reproducibility.
One last issue to note regarding file paths is that the syntax will be different in different operating systems, even for relative paths. The {here} package provides a simple way to wrap file paths so that they work across any operating system and integrates very smoothly into this project-oriented workflow.
Projects in practice
Using RStudio
To create a new project use the Create Project command (available on the Projects menu and on the global toolbar). You can create an RStudio project in a brand new directory or in an existing directory where you already have R code and data. This creates a project file (.Rproj) within the project directory that tells RStudio this is the root folder of your project.
There are several ways to open an existing project:
Using the Open Project command (available from both the Projects menu and the Projects toolbar) to browse for and select an existing project file (e.g. MyProject.Rproj).
Selecting a project from the list of most recently opened projects (also available from both the Projects menu and toolbar).
Double-clicking on the project file within the system file explorer (e.g. Windows Explorer, OSX Finder, etc.).
When opening an RStudio project, RStudio starts a fresh R session and sets the current working directory to the root of the project, so there is no need to call setwd() yourself. See why this is important in Reproducible Research.
See this RStudio tutorial for more detailed instructions.
Using the {usethis} package
The {usethis} R package provides a set of functions to create and manage R projects interactively. They do not need to be RStudio projects.
Useful functions to be run interactively in the R command line:
Create a new project with
usethis::create_project()and the absolute path where you want to create the project as the argument.Open an existing project with
usethis::proj_activate()and the absolute path of the project as argument.
Using other IDE
IDEs make it easy to work within this project-oriented workflow. You may prefer to use an IDE other than RStudio, such as VScode or Emacs + ESS, which come with their own project-related tools. In fact, these editors may be preferable when working on the remote cluster as they allow interacting with remote R sessions over SSH.
Standalone scripts
Each script in your R/ or scr/ folder should be standalone, meaning that any given script should not depend on global objects created in other scripts. Any objects created in one script that need to be used by other scripts should be explicitly written to a file somewhere within the project. Any other script that uses this file then needs to explicitly read the file.
For instance, if you save the data frame that you created in read-data.R inside the data/processed/ folder as an RDS file, you should then explicitly read these files from data/processed/ in any other script that uses these data frames.
Let’s see a very simple example. Let’s say we have a script read-data.R inside the R folder:
## R/read-data.R
data_raw <- read.csv("data/raw-data.csv")And a fit-model.R script:
## R/fit-model.R
model <- lm(y ~ x, data = data_raw)Script fit-model.R relies on the object data_raw loaded by script read-data.R in the global environment.
Instead, we can save the object created in the first script into a file and then load this file in the second script:
## R/read-data.R
data_raw <- read.csv("data/raw-data.csv")
saveRDS(data_raw, "data/processed/data_raw.rds")## R/fit-model.R
data_raw <- readRDS("data/processed/data_raw.rds")
model <- lm(y ~ x, data = data_raw)Analysis pipeline
A main script should be provided at the root of the project which runs all the required scripts in the right order to produce the final results of the analysis.
Instead of runing the code in each script “manually”, you should have a single script that orchestrates the whole process, thus making sure that the results across all the steps in the analysis are in-sync. Moreover, a master script can help a colleague looking at your analysis understand how to reproduce your results.
The name of this main script should be indicated in the README file of the project.
There are a few options for managing the pipeline of an analysis but a bare-bones approach can be to create a script that uses the source() function to evaluate each of your analysis scripts in the right order. This file should be at the root of the project folder.
## main-script.R at the root of your project
library(tidyverse)
library(mgcv)
source("R/read-data.R")
source("R/fit-models.R")
source("R/create-tables.R")
source("R/create-figures.R")There are more sophisticated and helpful ways to provide a master script. Check the targets R package and GNU Make.